Project name: ABCFEH; Project Leader: Tamás Hegedűs
Home inst.: MTA-SE Molecular Biophysics Research Group, the Hungarian Academy of Sciences, Budapest, Hungary and the Department of Biophysics and Radiation Biology, Semmelweis University, Budapest, Hungary
Cystic fibrosis is the most common monogenic disease in the Caucasian population, with high mortality. The fatality is associated with a lack of chloride efflux from the cell to the airway surface liquid that results in abnormally dense liquid. This phenomenon leads to dysfunctional cilia beating and impairs the clearing of dust and bacteria. Similar processes cause problems in the digestive system (gut, pancreas). On the other hand, in the skin there is no chloride resorption from sweat, so patients loose an important ion. The high loss of salt can easily be sensed, and had served for diagnosis, associated to witchcraft in the late fifteenth century and expressed in an Irish proverb: “Woe to that child who tastes salty when kissed on the forehead. He is bewitched and soon must die.”
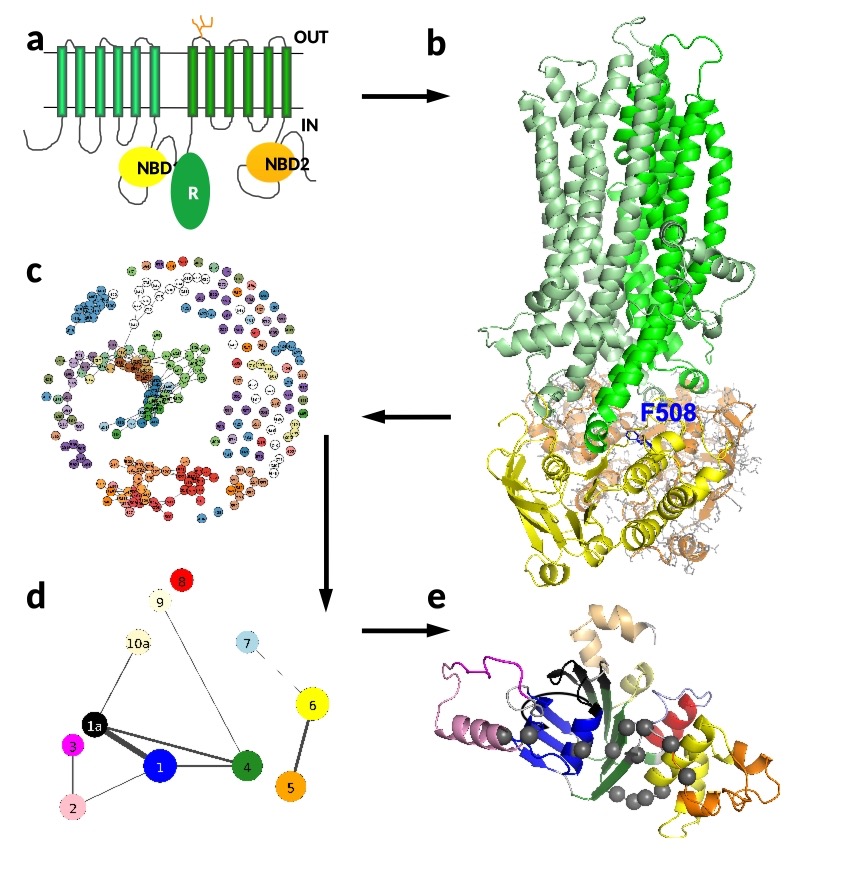
The cystic fibrosis disease is caused by mutations in the gene of the CFTR protein, which is a chloride channel in the cell membrane. When this protein is mutated, its structure, folding, and dynamics are affected. The main objective in the field is to design drugs to restore the normal structural and dynamic properties of CFTR and deliver this channel to the cell membrane. Knowing protein structure facilitates drug design and repair (a). The CFTR structure builds up from 6+6 transmembrane helices traversing the cell membrane, two intracellular nucleotide/ATP binding domains (NBD1 and NBD2). All the atoms of NBD2 are shown for demonstration, but the other parts have been simplified to ease the visualization (b). The most frequently mutated amino acid, F508, is highlighted by blue sticks. The deletion of this phenylalanine (ΔF508) disrupts not only the interaction between NBD1 (yellow) and transmembrane helices (green), but also effects the stability of NBD1.
Since experimental observation of atoms’ motions in proteins is extremely challenging or even impossible in most cases, we employ in silico methods to compute the movements based on physics to obtain a molecular level movie (http://www.hegelab.org/md.html). While it is easy to calculate the movement of one or two atoms, it is demanding for hundreds of thousands in a protein. This is caused not only by the high number of atoms, but also the complicated, long distance interactions between atoms, such as electrostatics. Therefore these so called molecular dynamics calculations need high performance computing resources (e.g. computers with a large number of CPUs and several high-end GPUs).
The analysis is also challenging, drawing on various statistical and network science methods. By probing the nature of communities in the amino acid network (c, d) it is possible to identify critical ones (e), which can enable the spread of potentially harmful mutations throughout the wider community of amino acids. These then become potential targets for drugs that can inhibit this spread.
Based on our results, the binding of thousands of molecules to that critical site can be tested in silico. The top hits, which include usually only tens of drug candidates, can be further tested using experimental methods. The experimentally effective and non-toxic molecules become leading molecules for further development and clinical tests. In summary, alloying knowledge of biology, physics, and mathematics can help in understanding the effect of mutations on protein structure and dynamics, in drug binding site identification, and potentially to develop therapy for diseases, such as cystic fibrosis.